Bacterial Genetics
This is based on New Zealand Qualification Authority Unit 12369 [1] entitled "Demonstrate knowledge of bacterial genetics."
1.1
Bacterial genomes
Modern molecular biology has defined the genome of any organism to be its hereditary information encoded in the chromosomal DNA. In haploid organisms (that include bacteria, viruses, archaea and mitochondria), the cell contains what was originally thought to be of a single,circular chromosome formation. Pulse-field electrophoresis(1989), provided clear evidence that some bacteria have single/multiple, circular/linear chromosomes, and single or multiple plasmids (explained later).
A bacterial genome is the complete gene complement contained on a single chromosome in a bacterium [2].
Bacterial DNA is single and circular having no nuclear membrane and no histones,some bacteria have one or more extra chromosomal elements which appear circular called plasmids,which mediates conjugation (Transfer of DNA by direct cell to cell contact) [3]
Prokaryotic chromosomes have shorter sequence of DNA. Typically, bacteria have a single circular chromosome with a single origin of replication[4].
An example of bacteria with more than typical chromosome is Agrobacterium tumefaciens which has one linear and one circular chromosome, and two circular plasmids! This bacteria is the causal agent of Crown Gall disease, a plant pathogen affecting grape vines, stone fruits and around 140 other economically important crops. The 'gall' or tumour is initiated by a small segment of 'transfer DNA' being incorporated into the plants genome. Details of 723 prokaryotic organisms [5] have been researched and published for reference. Further 'maps' of other bacterial genomes can be found at[6]
Bacteria reproduce by the process of binary fission. An explanation of this is available by consulting BACTERIAL CELL CYCLE AND CHROMOSOME REPLICATION: AN INTRODUCTION explains the nature of the bacterial genome including the structure and mode of replication of the bacterial chromosome. Replication of plasmids in gram-negative bacteria explains the nature of the bacterial genome including the structure and mode of replication of the plasmids.
A plasmid is an extra-chromosomal DNA molecule separate from the chromosomal DNA[7]. There are three general mechanisms of replication for bacterial plasmidsuch as theta type, strand displacement, and rolling circle.
[Power Point presentation - Describe: bacterial genome][8]
2.1
The bacterial gene contains a number of components and their key features are:
Operons: These are multiple genes of mRNA which are usually coded in single transcript.
Initiation codon: also known as start codon, encodes the amino acid methionine (fMet) in bacterial genes. The start codon is usually preceded by an untranslated region, 5' UTR.[9]
Open reading frame: It contains a sequence of bases that could encode a protein in them. These are found between the start and the stop codon.[10]
Ribosomal binding sites: When protein translation is initiated the sequence of mRNA is bounded by ribosome. It is known as ribosomal binding sites. [11]
Stop codon: Also known as termination codon. It is a mRNA which contains a nucleotide triplet that, basically, signals the termination of translation. [12]
Anatomy of Escherichia coli ribosome binding sites1 defines some of the components of a bacterial gene.
Typical structure of prokaryotic genes, which include bacterial gene, are illustrated with simple diagram.
This site has good information on transcription and translation, plus some other good basic information: http://en.wikipedia.org/wiki/Gene#Transcription
Thisc gives an information about the components of a bacterial gene[13]
[Power Point presentation - Describe: bacterial gene][14]
3.1 and 3.2
Transcription:[15]
DNA →RNA
On 5’ side of the gene protein (transcription factor) binds to the promoter side and RNA polymerase binds to the complex of transcription factor which results in the opening of the double helical DNA structure. This RNA polymerase reads one of the strands moving from 3’ to 5’. While moving through the DNA it assembles ribonuleotides into RNA strands following the base pairing rules. Therefore if C is present on DNA then G is inserted into RNA strand and if A is present on DNA then U will be inserted on RNA. The RNA forms in the 5’ to 3’ direction. As the process of transcription is complete, a transcript is released from the polymerase and soon the polymerase is released from DNA.
Translation:[16]
RNA → Protein
Translation involves various different processes which are as follows:
INITIATION: On the 5’side of gene the smaller subunit of the ribosome binds to the upstream site of the start of the message. It starts proceeding downwards until it reaches AUG which is a start codon. On reaching this point it joins with initiator tRNA and larger subunit. This tRNA binds to the P site.
ELONGATION: In this the aminoacyl-tRNA able to base pair with next codon on mRNA arrives at A site. This preceding amino acid covalently links through a peptide bond to the incoming amino acids. Finally the tRNA is released from the P site and the ribosome starts moving one codon downwards. This shifts tRNA (recently arrived) to the P site and opens A site for new aminoacyl-tRNA.
TERMINATION:
It is the end of the translation. It occurs when the ribosome reaches one or more stop codon. In this bacteria possess two different transcription termination codons.In Rho dependent one; a protein factor called Rho destabilize the interaction between the template and the mRNA, there by releasing a newly synthesized mRNA from the elongation complex. In Rho independent transcription termination; transcription stops when the newly formed RNA makes a hairpin loop followed by a run of uracil bases, which helps it to detach from the DNA template.[17]
This website outlines Gene Expression: Transcription for bacteria[18].
This website outlines Translationof messenger RNA to produce functional protein[19].
This gives diagrams and descriptions of the steps involved in transcription and translation [20]
[Power Point presentation - Describe: bacterial expression][21]
4.1
This is website outlines Control of Gene Expression in Bacteria
Gene expression is controlled through transcription[22]. Operon theory explains how the lac operon works
Lac operon is an example of gene expression control, it is necessary for the metabolism of lactose in E. coli and some enteric bacteria it also considered as an example for prokaryotic gene regulation.It produces a two part control mechanism.First control mechanism is the regulatory response to lactose,with the help of an intracellular regulatory protein called lactose repressor for assisting the production of beta galactosidase in the absence of lactose.The second control mechanism is associated with response to glucose in which catabolic activator protein(CAP) increases the production of galactosidase in the absence of glucose[23]
An example of an inducible operon: The lac operon in E. coli is to produce enzymes for breaking down of lactose. - it is normally off, but when a molecule called an inducer, lactose, is present, the operon turns on.[24]
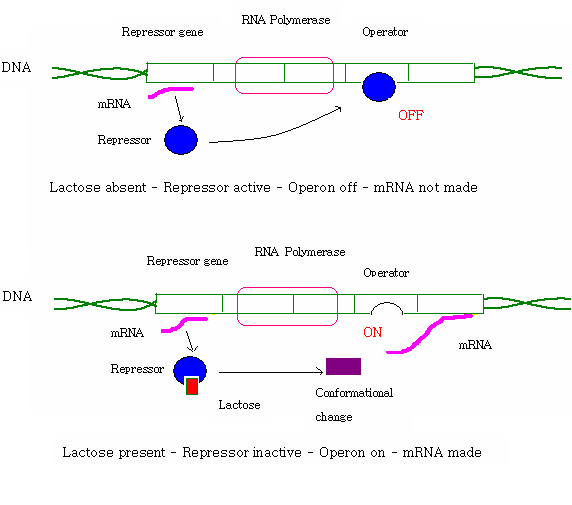
[Power Point presentation - Describe: gene control][25]
5.1 and 5.2
Types of mutations[26]
Point Mutation: A simple change in one base of the gene sequence, the most common.
Deletion: Deletions of just one single base, affects a large number of genes on the chromosome.
Insertion: Addition of one or more bases, affects the protein structure significantly.
Silent mutation: Has no effect on a protein.
substitution: exchanges one base for another.It changes a codon to one that encodes different amino acid results a small change in the protein.[27]
Mutations in 16S rRNA and Ribosomal Protein S5 Associated with high-Level spectinomycin resistance in Pasteurella multocida gives an example of a range of mutations that can occur from alterations to bacterial genes. gives an example of the mode of action of mutagens effect on bacterial genes.
A mutation occurs when a DNA gene is damaged or changed in such a way as to the nucleotide sequence of the genetic material of an organism[28].
Mutagens are the chemical or physical agents that cause changes in the genetic information of an organism and there by increasing the frequency of mutation.Some of them act as a base analogs, and get inserted in to the DNA strand during the placement of substrates.Some of them react with DNA to produce a structural change which leads to the miscopying of the template strand when the DNA is replicated.Some work indirectly by causing the cells to synthesize chemicals that have the direct mutagenic effect.
[29]
Types of Mutagens[30]
Chemical mutagens: altering structure of bases.
Radiation: breaking in one or both strands, damaging of bases, or cross-linking of DNA to itself or proteins.
UV: producing pyrimidine dimers and blocking transcription and DNA replication.
A mutagen[31] is a physical or chemical agentthat changes the genetic information of an organism and thus increases the frequency of mutations.
[Power Point presentation - Describe: gene alteration][32]
6.1, 6.2 and 6.3
Reproduction of bacteria occurs by fission and the exchange of hereditary characteristics, called horizontal gene transfer.There are three ways for the exchange of DNA between bacteria, conjugation, transduction and natural transformation. In conjugation a piece of DNA is copied from one bacterium and transferred to another by a temporary connection called conjugation pilus. In transduction transfer of DNA takes place with the aid of a bacteriophage.In transformation DNA is located outside the cell and is fragmented and imported in to the cell by recombination.[33]
Gene transfer is a big part of bacterial evolution. Bacterial Gene Transfer by Natural Genetic Transformation in the Environment outlines the major methods.EXCHANGE OF GENETIC INFORMATIONdescribes the structure and properties of transposons and insertion sequences.Antibiotic resistance gives examples of the effects of genetic material transfer between bacteria on the dissemination of drug resistance elements.
Simple diagram of gene transfer that makes easy to spread out drug resistance.
Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments.
Transposons and Insertion sequences
Transposons called "jumping genes" are pieces of DNA which can move around to different locations within the genome. Sometimes transposons change the amount of DNA in the genome and cause damage to the DNA.[34] They can facilitate the transfer of genes, such as antibiotic-resistance genes, from the chromosome of a bacterium to a plasmid.[35]
Insertion sequences(IS) are small transposons carrying only the genetic functions involved in transposition. They have at their ends repeated sequences encoding an enzyme called transposase which enables the IS to move.[36]
Major methods of gene transfer between bacteria


[Power Point presentation - Describe: gene transfer][37]